🗨 About Me
I am a first-year PhD student in the Computational Biology and Bioinformatics (CBB) program at the University of Southern California. My research lies at the intersection of AI and biology, where I design computational approaches to accelerate discoveries in synthetic biology, drug discovery, and molecular interaction. Specifically, I primarily focus on those areas: (1) AI driven drug discovery [SMARTBind, Apo2Mol]; (2) Biological foundation model [Tabula, ProTrek, SaProtHub, Nullsettes]; (3) Machine learning enabled protein evolution [Sequence Display]. I am also interested in bioinformatics tool and database development [SICER 2.0, gmx_mmpbsa_py, HNOXPred].
Before starting my PhD, I was very fortunate to work with and learn from inspiring mentors and collaborators across these fields, you can find them in the experience panel.
📖 Educations
- 2025 - current, PhD student, Computational Biology and Bioinformatics. University of Southern California. Los Angeles, CA
- 2022 - 2024, Master of Science in Engineering, Computer Science. Johns Hopkins University. Baltimore, MD
- 2018 - 2022, Bachelor of Science, Computer Science. Wenzhou-Kean University. Wenzhou, China
📰 News
- 2026.02: “Sequence Display: Generating Large-Scale Sequence–Activity Datasets to Advance Universal Protein Evolution” is accepted in principle by Nature Biotechnology!
- 2026.01: “Small Molecule Approach to RNA Targeting Binder Discovery (SMARTBind) Using Deep Learning Without Structural Input” is presented at UF Health Cancer Institute Annual Research Showcase 2026, see our poster.
- 2025.11: “Apo2Mol: 3D Molecule Generation via Dynamic Pocket-Aware Diffusion Models” is accepted by AAAI 2026, see our poster.
- 2025.10: “Engineering Unnatural Cells with a 21st Amino Acid as a Living Epigenetic Sensor” is on Nature Communications!
- 2025.09: “Small Molecule Approach to RNA Targeting Binder Discovery (SMARTBind) Using Deep Learning Without Structural Input” is released on bioRxiv.
Earlier News (Click to Expand)
- 2025.09: “Tabula: A Tabular Self-Supervised Foundation Model for Single-Cell Transcriptomics” is accepted by NeurIPS 2025!
- 2025.09: “Biosynthesis of Unnatural Cyclodipeptides through Genetic Code Expansion and Cyclodipeptide Synthase Evolution” is on Journal of the American Chemical Society!
- 2025.08: “Evaluating DNA function understanding in genomic language models using evolutionarily implausible sequences” (follow-up work from the GenBio workshop) is released on arXiv.
- 2025.08: “SaprotHub: Democratizing Protein Language Model Training, Sharing and Collaboration for the Biology Community” is accepted by Nature Biotechnology!
- 2025.07: “A tri-modal protein language model enables advanced protein searches” is accepted by Nature Biotechnology!
- 2025.07: “Predicting function of evolutionarily implausible DNA sequences” is presented at Q-BIO 2025 Conference: Emergent Orders in Living Systems Across Scales, see our poster.
- 2025.06: “Sequence Display-Enabled Machine Learning for Protein Evolution” is presented at 2025 Synthetic Biology: Engineering, Evolution, & Design, see our poster.
- 2025.06: “Predicting function of evolutionarily implausible DNA sequences” is accepted by ICML 2025 Generative AI and Biology Workshop!
- 2025.04: I will be joining the PhD program in Computational Biology and Bioinformatics at USC QCB, looking forward to the journey.
- 2025.01: “Toward a privacy-preserving predictive foundation model of single-cell transcriptomics with federated learning and tabular modeling” is released on bioRxiv, see our post.
📝 Selected Publications
2026
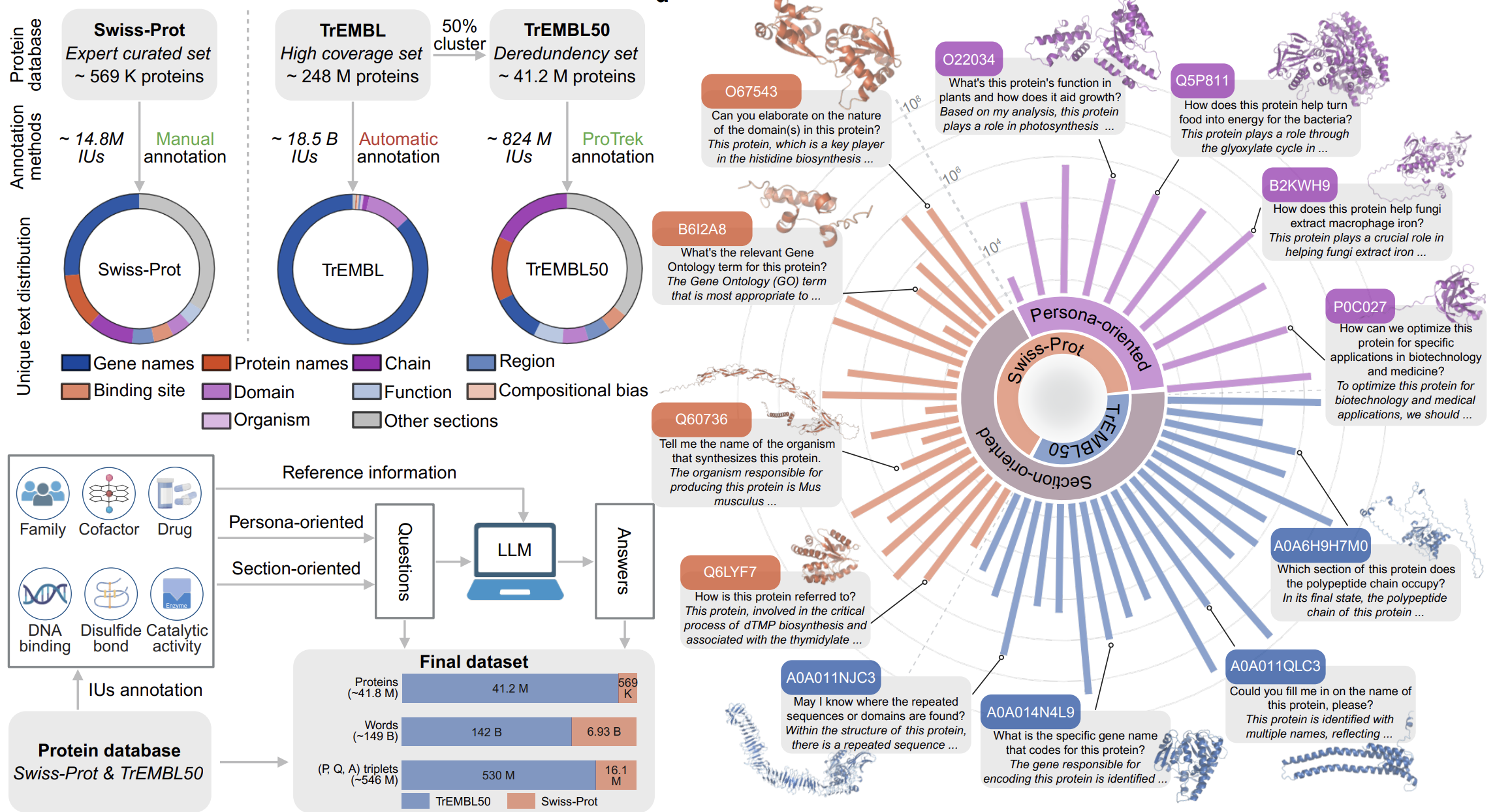
Decoding the Molecular Language of Proteins with Evolla
Xibin Zhou †, Chenchen Han †, Yinqi Zhang †, Huan Du †, Jiayuan Tian †, Jin Su ‡, Renju Liu ‡, Kai Zhuang ‡, Shiyu Jiang ‡, Anthony Gitter, …, Zongze Zhao, Yang Liu, Hongyuan Lu, Fajie Yuan. bioRxiv, 2026. (Under review in Nature Research Journal)
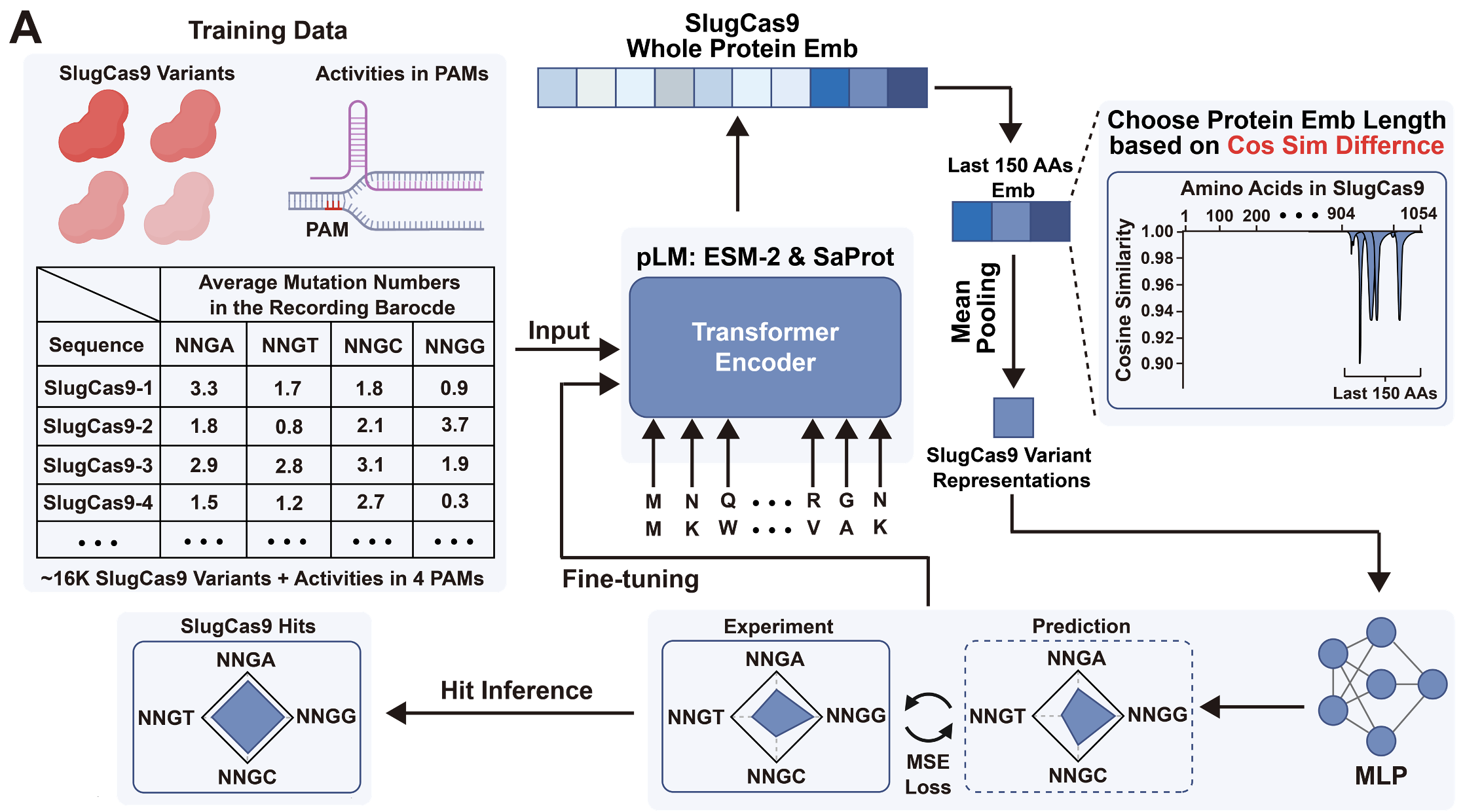
Linqi Cheng †, Xinzhe Zheng †, Shiyu Jiang †, Hu Y, Liu Y, Yang K, Rui J, Ding H, Zhang M, Yuan T, Ye H, Li C, Kevin K. Yang, Xiongyi Huang, Han Xiao. Nature Biotechnology, 2026. (Accepted in principle)
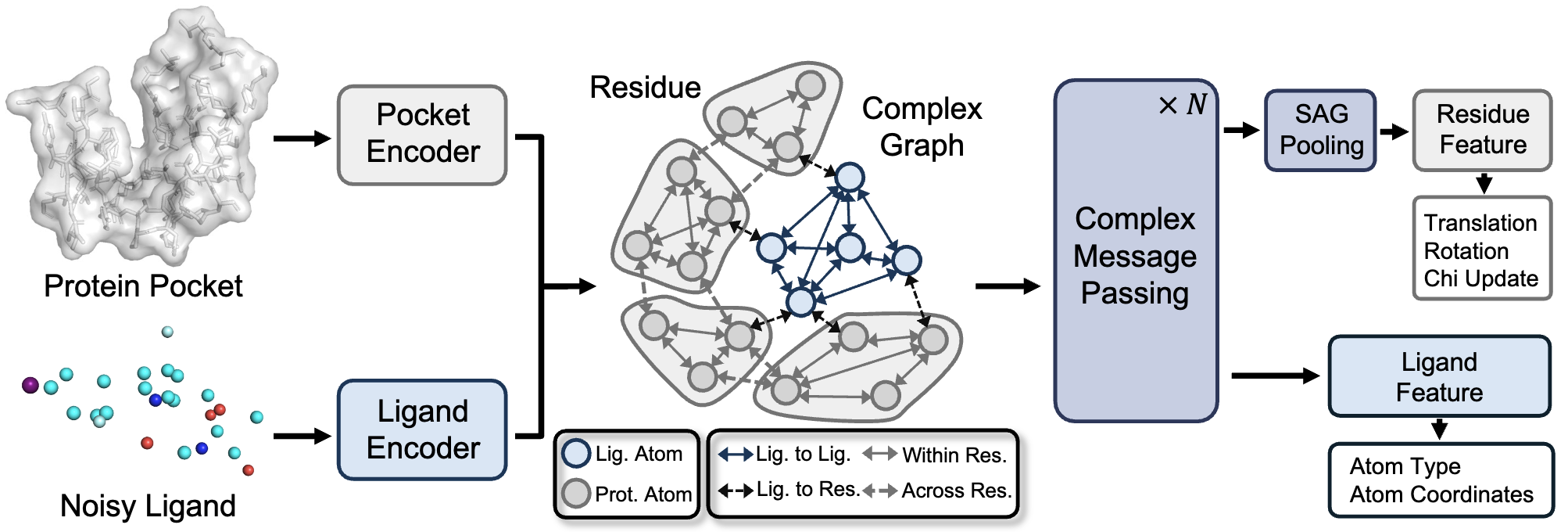
Apo2Mol: 3D Molecule Generation via Dynamic Pocket-Aware Diffusion Models
Xinzhe Zheng, Shiyu Jiang, Gustavo Seabra, Chenglong Li, Yanjun Li. AAAI (poster), 2026.
2025
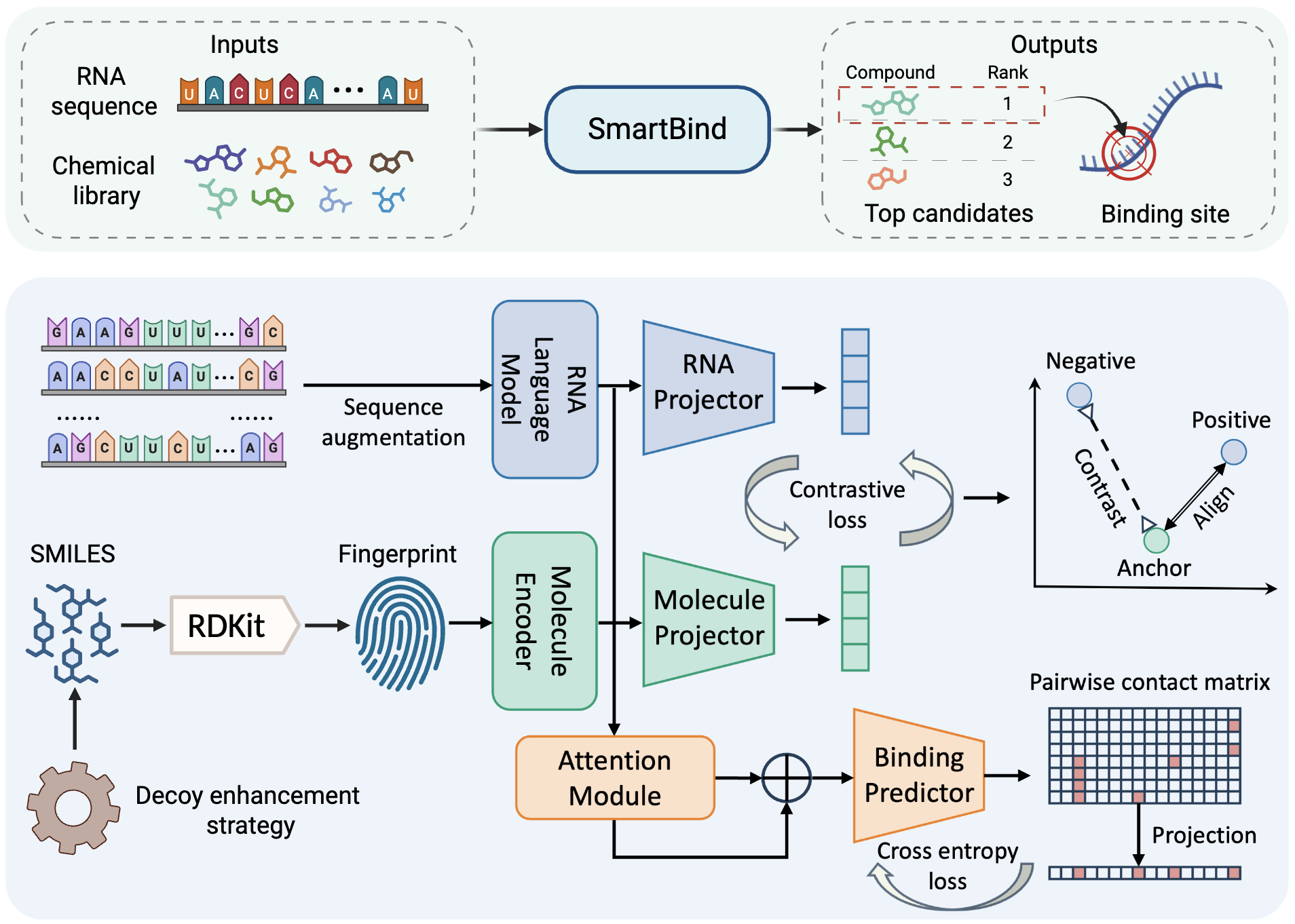
Shiyu Jiang †, Amirhossein Taghavi †, Tenghui Wang, Samantha M. Meyer, Jessica L. Childs-Disney, Chenglong Li, Mattew D. Disney, Yanjun Li. bioRxiv, 2025. (Under major revision in Nature Research Journal)
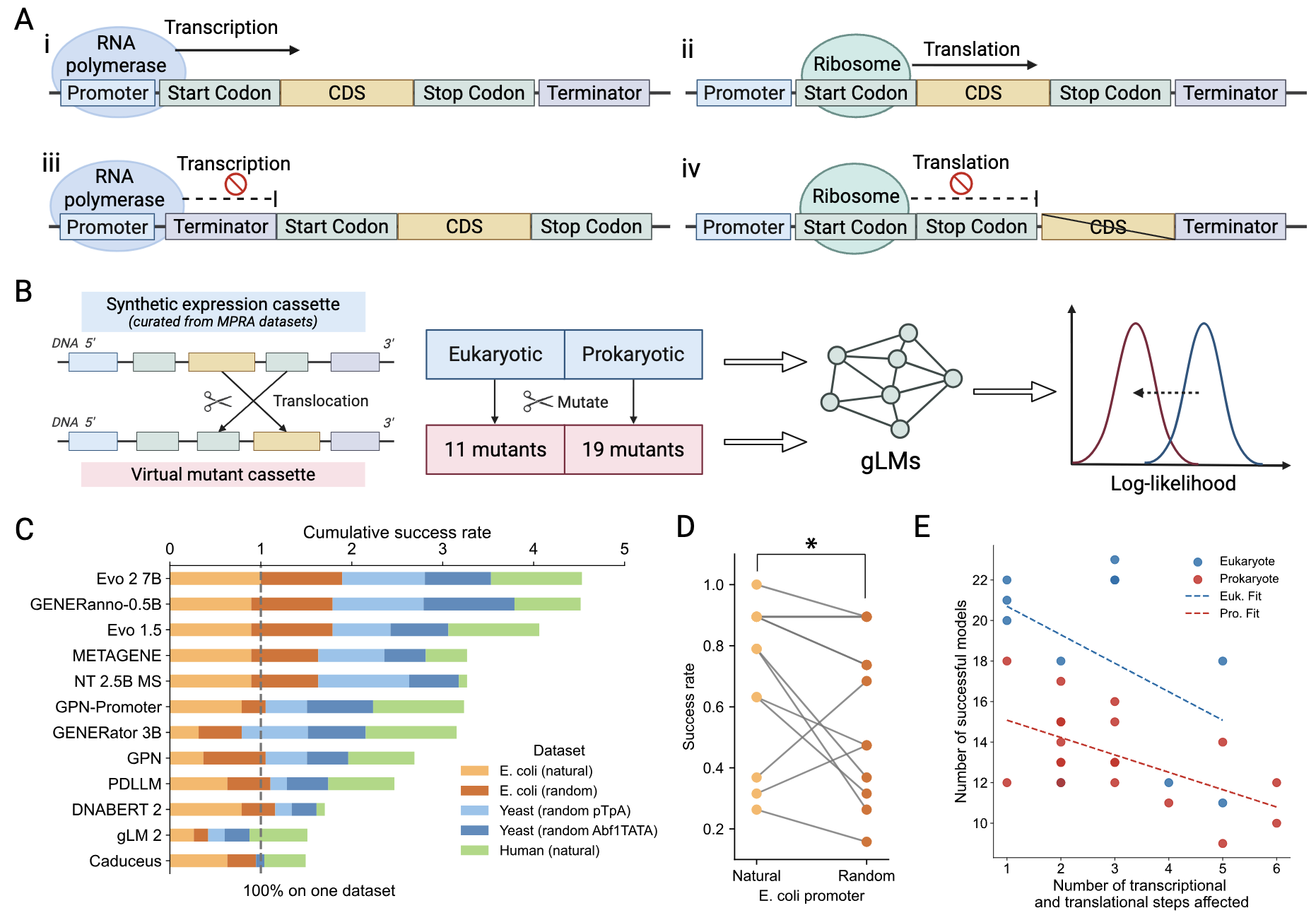
Shiyu Jiang, Xuyin Liu, Jerry Zitong Wang. arXiv, 2025. (Under review in Research Journal)
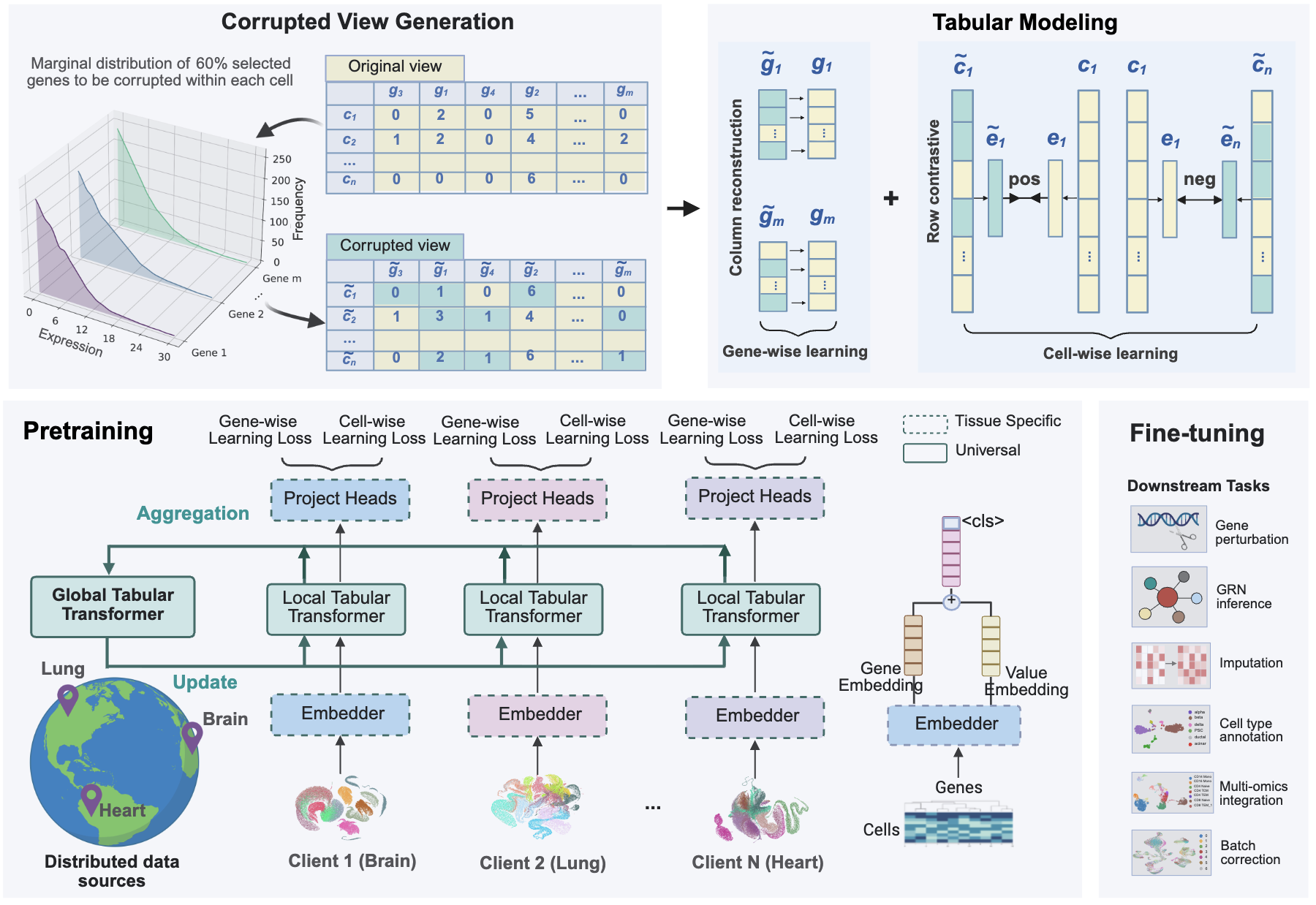
Tabula: A Tabular Self-Supervised Foundation Model for Single-Cell Transcriptomics
Jiayuan Ding †, Jianhui Lin †, Shiyu Jiang †, Yixin Wang, Ziyang Mao, Zhaoyu Fang, Jiliang Tang, Min Li, Xiaojie Qiu. NeurIPS (poster), 2025.
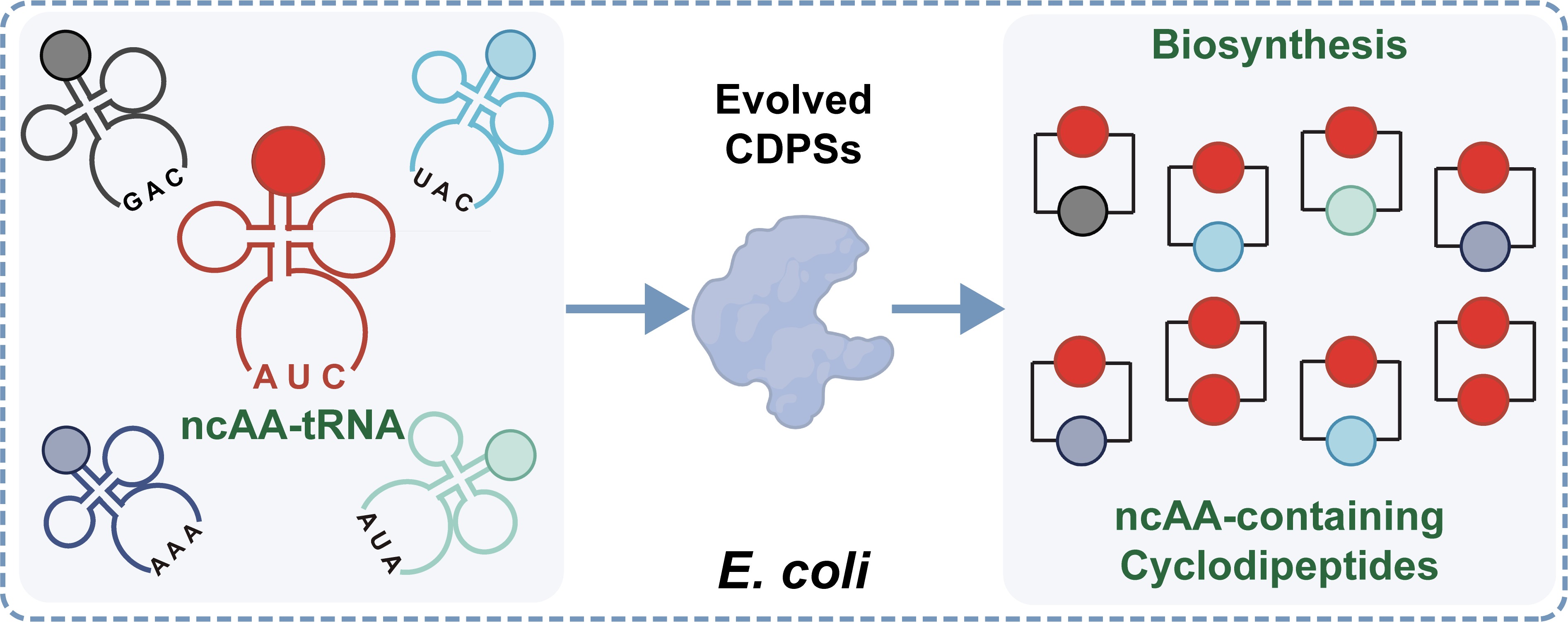
Hu Y †, Cheng L †, Liu Y, Liu R, Jiang S, Yuan T, Wang Y, Ye H, Xiao H. Journal of the American Chemical Society, 2025.
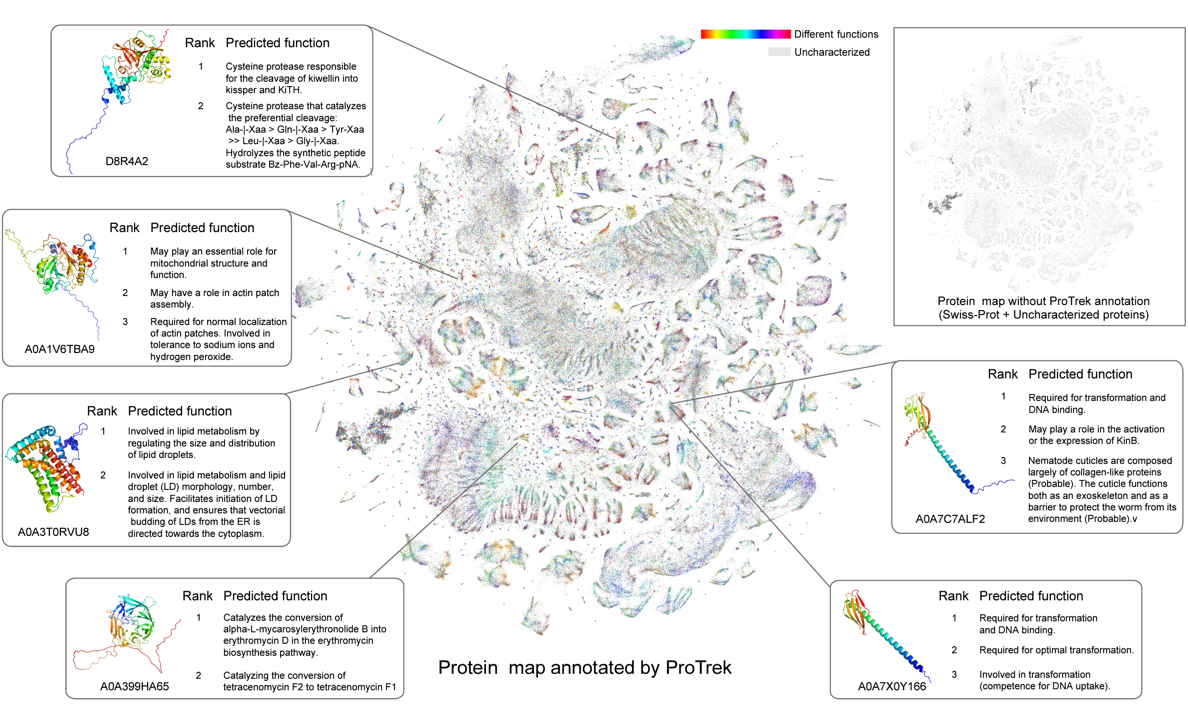
A tri-modal protein language model enables advanced protein searches
Jin Su †, Yan He †, Shiyang You †, Shiyu Jiang, Xibin Zhou, Xuting Zhang, Yuxuan Wang, Xining Su, Igor Tolstoy, Xing Chang, Hongyuan Lu, Fajie Yuan. Nature Biotechnology, 2025.
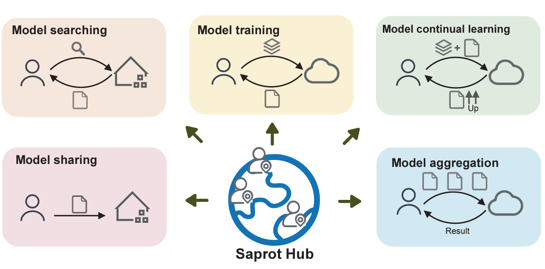
Jin Su, Zhikai Li, Tianli Tao, Chenchen Han, Yan He, Fengyuan Dai, Qingyan Yuan, Yuan Gao, Tong Si, Xuting Zhang, Yuyang Zhou, Junjie Shan, Xibin Zhou, Xing Chang, Shiyu Jiang, Dacheng Ma, The OPMC, Martin Steinegger, Sergey Ovchinnikov, Fajie Yuan. Nature Biotechnology, 2025.
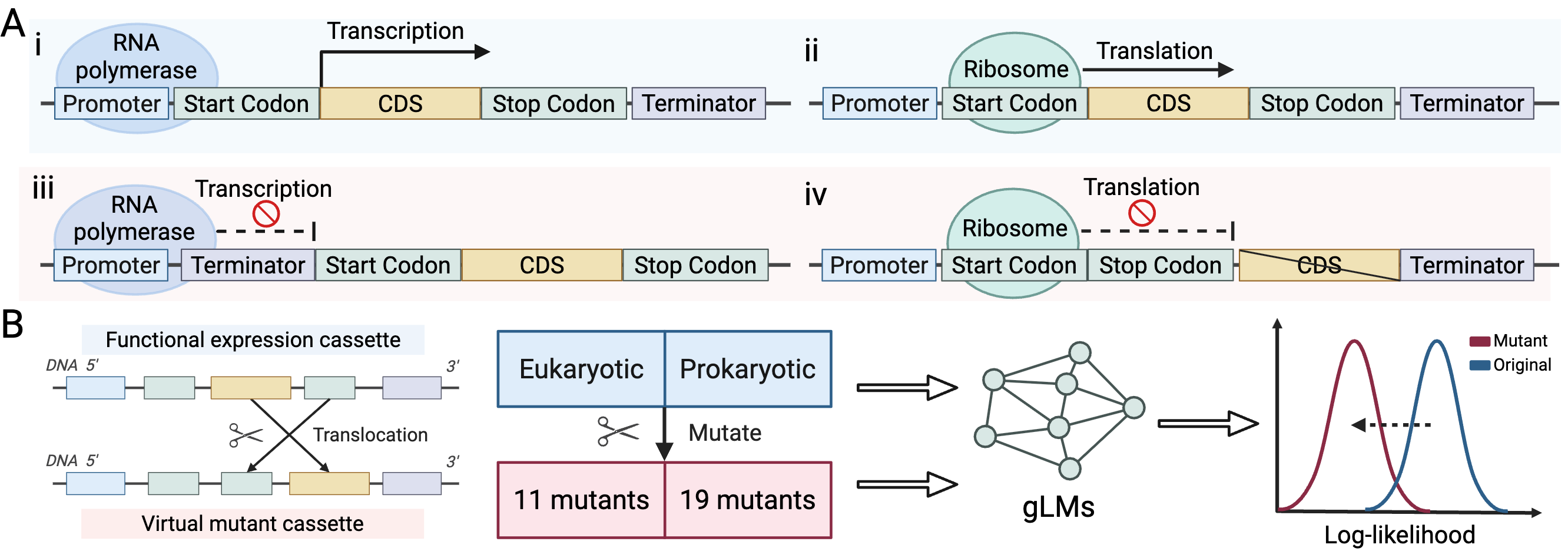
Predicting function of evolutionarily implausible DNA sequences
Shiyu Jiang, Xuyin Liu, Jerry Zitong Wang. ICML 2025 Generative AI and Biology Workshop, 2025.
2024
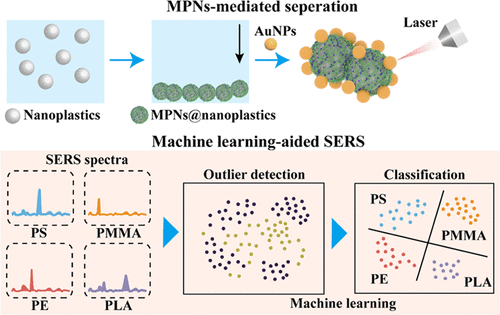
Haoxin Ye, Shiyu Jiang, Yan Yan, Bin Zhao, Edward R Grant, David D Kitts, Rickey Y Yada, Anubhav Pratap-Singh, Alberto Baldelli, Tianxi Yang. ACS Nano, 2024.
2023
Simulating Disease Spread During Disaster Scenarios
Shiyu Jiang, Heejoong Kim, Fabio Henrique Tanaka, Claus Aranha, Anna Bogdanova, Kimia Ghobadi, Anton Dahbura. The International Conference on Artificial Life, 2023.
2022
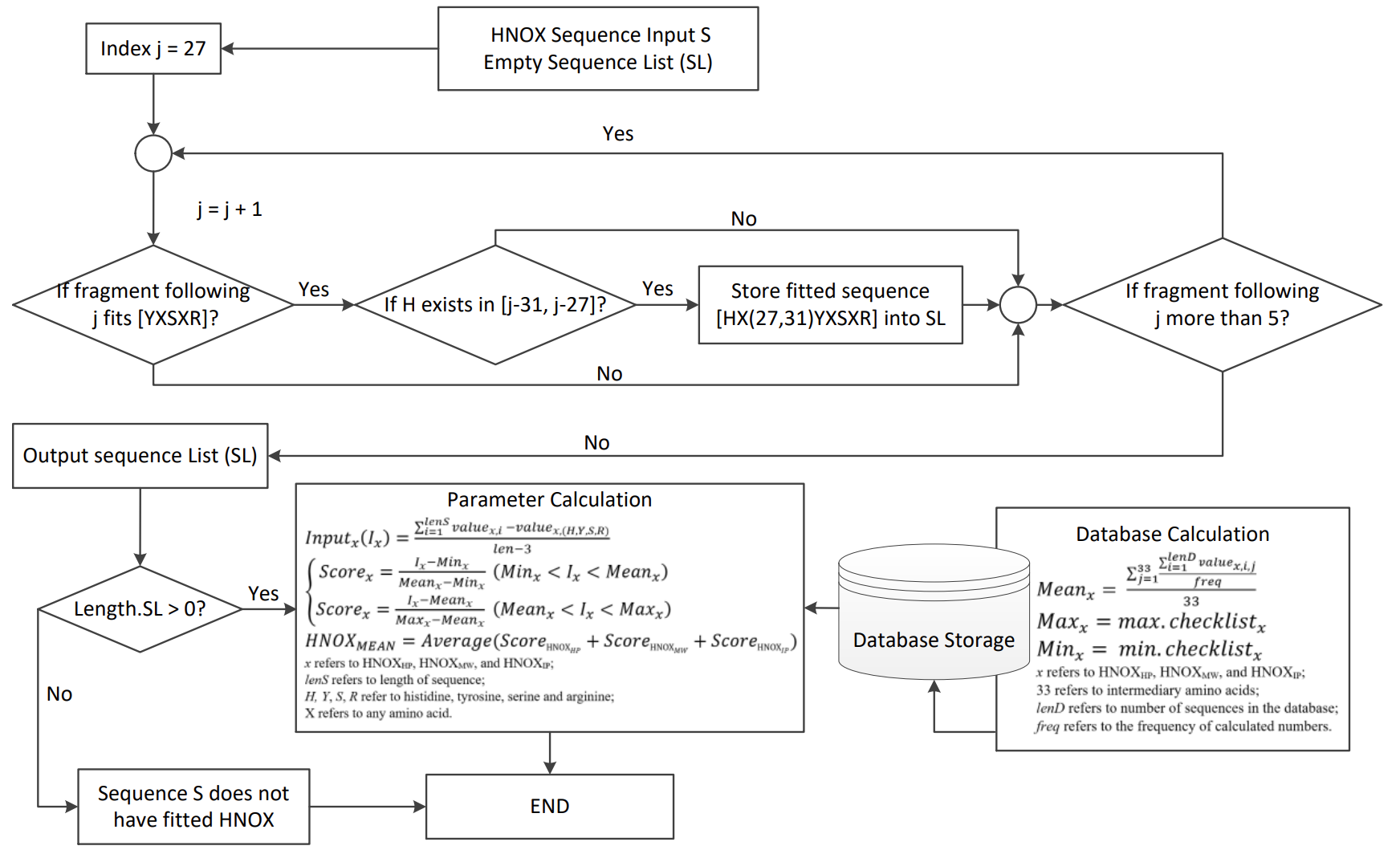
HNOXPred: a web tool for the prediction of gas-sensing H-NOX proteins from amino acid sequence
Shiyu Jiang, Hemn Barzan Abdalla, Chuyun Bi, Yi Zhu, Xuechen Tian, Yixin Yang, Aloysius Wong. Bioinformatics, 2022.
🧑💻 Experience
-
2025.09 - Present
Graduate Research Assistant
University of Southern California, Department of Quantitative and Computational Biology
-
2024.08 - 2025.06
Research Associate
Development and evaluation of protein/genomic language model | Advisor: Prof. Fajie Yuan & Dr. Zitong Jerry Wang
Westlake University, School of Engineering & Center for Interdisciplinary Studies, School of Science
-
2023 - 2025
Remote Research Assistant
Protein language model driven protein evolution with sequence display | Advisor: Prof. Han Xiao
Rice University, Department of Chemistry
-
2023 - 2025
Remote Research Assistant
RNA-small molecule drug discovery and protein-molecule generation | Advisor: Prof. Yanjun Li & Prof. Matthew D. Disney
University of Florida, College of Pharmacy & UF Scripps Institute, Department of Chemistry
-
2023 - 2025
Remote Research Assistant
Foundation model for single-cell transcriptomics | Advisor: Prof. Xiaojie Qiu
Stanford University, Department of Genetics
-
2024.01 - 2024.07
Lab Specialist
ChIP-Seq peak calling tool | Advisor: Prof. Chongzhi Zang
University of Virginia, Department of Genome Sciences
-
2021 - 2022
Undergraduate Research Assistant
Bioinformatics webtool development | Advisor: Prof. Aloysius Wong
Wenzhou Kean University, Department of Biology
📝 Service
- Journal reviewer: IEEE Transactions on Computational Biology and Bioinformatics
- Conference reviewer: AAAI 2026
🌎 Miscellaneous
Outside of work, you’ll often find me at the gym, playing soccer, road cycling, or go hiking. I also enjoy playing table tennis and the piano occasionally.









